Jason Evans is giving me a tour of the new NCI Program for Natural Products Discovery (NPNPD) facility when he pauses to look at a sample-handling robot under repair, its mechanical insides temporarily disemboweled. I ask if he has an electrical engineering background, and he laughs. Evans is a scientific programmer, but for him and his colleagues, that’s beside the point.
“It’s baptism by fire here,” he says, explaining that everyone in the Natural Products Support Group, a team from the Frederick National Laboratory that works in tandem with the NPNPD, learns to repair the robots to some extent.
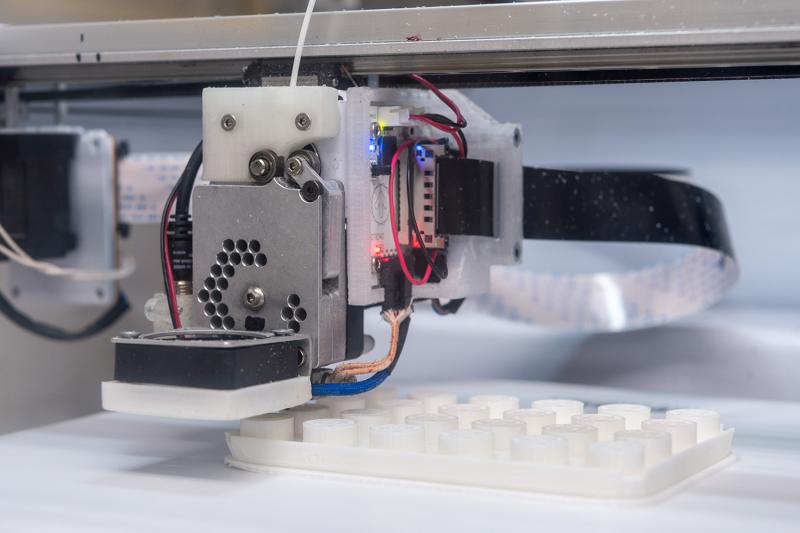
Evans and his colleagues oversee an ecosystem of automated machines that prepare, move, weigh, store, and ultimately distribute samples to dozens of academic labs around the globe. One particularly impressive machine dashes back and forth across a 40-foot-long cold storage unit, retrieving samples from 15-foot-tall racks and then carefully transferring them through a small chute to a waiting robotic arm. Another draws liquid into a set of pipettes and transfers it to pre-labeled tubes, which are then whisked away to a different robot. Much of the material being used in this process, from pipette tips to tube caps, has been 3D printed in-house—a substantial cost-saving measure.
This array of technological marvels is helping rekindle the search for drug candidates sourced from natural products. Though roughly half of all existing cancer drugs (such as Eribulin, which originated from a marine sponge) are derived from plants, animals, and fungi, such products have become less popular with researchers in recent years, in part because they are difficult to test with modern high-throughput screening techniques.
To help reinvigorate natural products research, NPNPD’s interdisciplinary team of biologists, chemists, programmers, and technicians is building an enormous library of natural products fractions (samples that have been purified and are ready to be tested) that should make the drug discovery process more efficient and less error-prone.
Automation is a key part of constructing the fractionated library. Without it, NPNPD’s team would need to be much, much larger to replicate the work done by the efficient robots. But an equally important part of the project is data analytics and visualization.
Every machine in the gleaming facility is connected to a central website that collects data about the robots’ tasks for record keeping and analysis. In addition, NPNPD’s researchers are generating an enormous amount of data as they build their sample library that, properly organized, could help scientists identify the most interesting compounds and extracts and eliminate redundant testing on products that show similar traits.
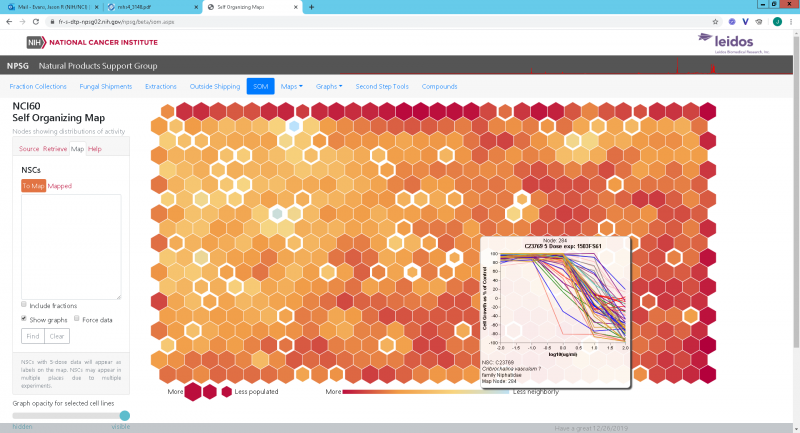
Evans, when he isn’t checking on a handling robot or testing a 3D printer, is using a combination of biology, computer science, and statistics known as bioinformatics to organize NPNPD’s data. With other members of the Natural Products Support Group, he helped create a self-organizing map—a machine learning system that organizes samples based on their “topological” characteristics.
“The attempt is to organize the data that we get into discreet units of activity so that we can tell whether we’ve seen this pattern before and whether any work has been done on it,” Evans said. “It’s a way to cut down on the amount of time a chemist would spend on the project by providing preliminary information or [helping them] decide that [a certain lead] isn’t going anywhere so they can move on to something more interesting.”
Hexagonal nodes sprawl across the map, ranging in hue from pale yellow to deep orange and red. The colors represent the degree of similarity between neighboring hexagons, while their sizes correlate with the quantity of samples that exist in each node. Users can click on each hexagon to see the profiles in a given node, including the structures of the compounds and the taxonomies of the extracts—with a few caveats. There are limits to the information that the group will make publicly available, in part because they don’t want to expose vulnerable regions or species to potential piracy.
“The gold medal would be to find a pure compound that shows the same activity as an extract and say, okay, it’s probably a class of these compounds that’s causing the activity if they are grouped together, so maybe this organism produces something like it,” Evans said. “Then you can jump to the end with some confidence and have more-targeted experimentation.”
Evans and his colleagues hope to make a version of the map publicly available as a tool to help collaborators prioritize their workloads. In the meantime, the team will keep the automated facility humming along so that the extract library keeps growing. To date, NPNPD has purified about 330,000 fractions, which puts them a third of the way to their goal of 1 million.
It’s an expansive undertaking that requires the right people, the right tools, and a certain “do-what-it-takes” attitude when something breaks down. Fortunately, that’s just what this group has.