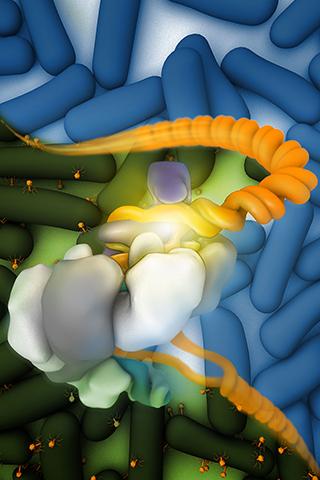
To get a closer look at one CRISPR complex, researchers from NCI’s Center for Cancer Research and their collaborators recently put it “on ice” with cryo-electron microscopy (cryo-EM), creating highly detailed images that show its biological structures in multiple states at a molecular level.
Scientists have known about CRISPR (clustered regularly interspaced short palindromic repeat) complexes since the early 2000s. In nature, they act as a sort of immune system for many types of bacteria: through a process that involves removing and processing sequences of an invading virus’ DNA, bacteria produce CRISPR RNA strands that join with their Cas proteins to recognize and degrade the target sequences in the virus, eventually neutralizing the invader and—on a large scale—halting future invasions.
The CRISPR-Cas complexes’ “sequence-snipping” function and their dependence on an initial input make it possible to program them to modify a subject’s DNA, something scientists recognized five years ago when they began using them for genome editing. Since then, the use of CRISPRs has soared, and their popularity for modifying cell lines and experimental models continues to grow.
Naturally, that utility also underscores the need for a clear understanding of the complexes’ structures and functions at a molecular level, along with the potential for using emerging methods in cryo-EM to meet that need.
In their recent study reported in the journal Cell, the researchers investigated the structural states of CRISPR system yersinia (Csy)—a CRISPR-Cas complex found in the bacteria Pseudomonas aeruginosa—before, during, and after binding to viral double-stranded DNA (dsDNA) and three viral Csy inhibitors.
The study, which used the Krios microscope at the Center for Molecular Microscopy in the Frederick National Laboratory for Cancer Research, was led by Sriram Subramaniam, Ph.D., of the Center for Cancer Research, in collaboration with the laboratory of Dinshaw Patel, Ph.D., at the Memorial Sloan Kettering Cancer Center in New York.
The analysis of the 3-D images provided multiple “new insights into how a CRISPR complex recognizes DNA destined for degradation and how selected inhibitors block DNA binding,” said Subramaniam, who is also director of the National Cryo-Electron Microscopy Facility at the Frederick National Laboratory.
Among the team’s findings was the discovery that, when bound to target dsDNA, Csy’s CRISPR RNA forms a hook that holds the DNA strand in place during the degradation process. The DNA-bound Csy complex also stretches along its axis, which Subramaniam believes may allow greater access to nucleases—enzymes that cleave DNA.
In addition, the team was able to determine the various ways the Csy inhibitors attached themselves to Csy’s CRISPR RNA and prevented the complex from binding to dsDNA or forming its hook.
Collectively, the results make an important addition to the field of CRISPR research. CRISPR-Cas complexes are highly diverse—there are 19 Cas protein subtypes, and, while many of them have a DNA degradation function as part of a CRISPR-Cas complex, they behave in different ways and have vastly different structures. Thus, the ability to visualize such differences in great detail enhances scientific understanding of CRISPR structural biology and may improve gene editing techniques.
“Our results help define the types of interactions involved in DNA binding and could be helpful for future biotechnology applications,” Subramaniam said.
According to the researchers, the findings also illustrate cryo-EM’s practicality for studying CRISPR-Cas complexes. Cryo-EM was a runner-up for Science magazine’s 2017 Breakthrough of the Year, and it brings more clarity and flexibility than traditional microscopy modalities like X-ray crystallography. Whereas crystallographic imaging relies on confined 3-D crystals, cryo-EM employs a thin aqueous buffer that is flash-frozen in liquid ethane, allowing for a less restrictive view with fewer visual artifacts.
“We anticipate we will be able to extend these types of analyses to study other classes of CRISPR complexes, especially those involved in gene editing,” Subramaniam said.