Editor’s note: A version of this story appeared on the Frederick National Laboratory website.
A mutated, cancer-causing protein twisted and bent across the computer screen. As Ruth Nussinov, Ph.D., and her team watched the simulation, two things quickly became evident. First, it was clear how the mutation paved the way for cancers to form. Second, the twisting and bending created a pocket—a gap in the protein’s proverbial armor—no one had seen before.
Nussinov’s team at Frederick National Laboratory for Cancer Research (FNL) has illuminated through computational simulations how mutated versions of that protein, mTOR, contribute to cancer at a molecular level. Their discovery of the heretofore-unknown pocket also offers an opportunity to target mutant mTOR to better treat cancer. Their findings appear in the Journal of Chemical Information and Modeling.
mTOR, which is short for “mechanistic target of rapamycin,” has roles in biological processes ranging from metabolism to aging. Crucially, it helps regulate essential survival functions in our cells. But some mutations allow it to become overactive. In those scenarios, cells multiply too fast, live too long, and are harder to kill—hallmarks of cancer.
It’s unsurprising that mutant mTOR has been implicated in melanoma and in malignant tumors in no less than six different organs. The protein is a high-profile candidate for cancer treatment, yet few drugs exist that specifically target its mutant forms.
mTOR’s structure—the configuration of its molecules, including folds, bends, and pockets—tells scientists how it functions and interacts with other proteins, both in cancer and normally. This information can guide efforts to develop new drugs, yet there’s still much unknown.
“This gap in understanding motivated us to investigate mTOR’s molecular architecture, as exploring these structural details could lead to more targeted and effective therapeutic strategies for cancer treatment,” said Nussinov, who leads the Computational Structural Biology Program, an FNL laboratory that’s embedded in the National Cancer Institute’s Center for Cancer Research.
Protein Unfolds as Part of Cancer Contribution
Nussinov’s team selected several cancer-causing mTOR mutants, then put them through molecular dynamics simulations, computer models that use existing chemical and biological data to predict how proteins behave under given conditions. The method let them render versions of mTOR with individual mutations and see how the mutations changed the protein’s structure, if at all.
“This approach also allows us to probe the effects of mutations or changes in environmental conditions on mTOR’s function, which would be difficult to observe directly with experimental methods,” Nussinov said. “Computational MD gives us a powerful tool to gain a deeper understanding of mTOR’s behavior in a way that complements experimental work and helps guide further investigation.”
The simulations revealed that most of the mutations shift mTOR’s structure in a way that favors cancer-causing conditions. In these mutants, a portion of mTOR unfolds such that other proteins can more frequently interact with it to trigger cells to multiply, overstay their life spans, and form tumors. Two other mutations stopped short of doing so but created conditions that make the shift likely.
“That was both exciting and surprising, as it suggested that small, previously overlooked changes in mTOR’s structure could have significant functional consequences,” Nussinov said, adding that some of the shifts aligned with known cancer-causing mutations. The finding underscores the relationship between mTOR’s structure and its role in cancer.
Pocket Suggests Treatment Is Possible
Potential treatments could aim to re-fold mTOR mutants or block their interactions while unfolded.
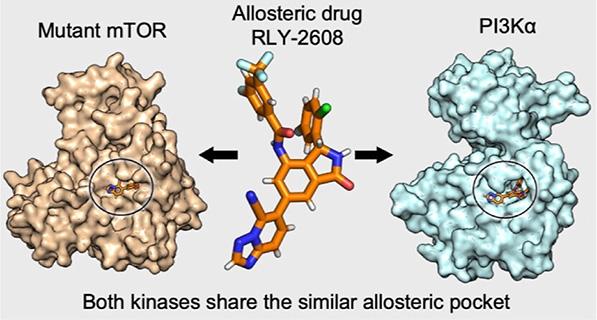
The new pocket the team discovered has potential for that. The same mutations that contort mTOR into its cancer-driving shape also create the pocket in its structure, and the simulations suggest at least two existing drugs can hit it.
The compounds, RLY-2608 and STX-478, were designed to disable a protein that commonly interacts with PI3Ka (a critical drug target for cancer), but they cleanly fit into the mutant mTOR’s pocket, the team found. Equally valuable, the drugs fit better with mutant mTOR than normal mTOR, suggesting that future treatments targeting the pocket may be better at zeroing in on the cancer cells with the mutants and less harmful to cancer patients’ healthy cells.
“Our discovery opens up new avenues for designing therapeutic strategies targeting mTOR,” Nussinov said.
Cautious Optimism
While Nussinov and her team feel enthusiasm over the outcome of the study, they’re balancing it with a healthy dose of realism. The work was done on the computer, and like any experimental method, molecular dynamics has limitations.
The findings are cause for optimism and testify to molecular dynamics’ power and utility, Nussinov said. She added, however, that they need to be validated in laboratory and microscopy studies so scientists can be certain the simulations align with reality.
“By integrating both, we expect to gain a deeper understanding of mTOR’s druggable potential,” she said. “Looking ahead, we are excited about collaborating to integrate our MD simulations with experimental data.”
Samuel Lopez leads the editorial team in Scientific Publications, Graphics & Media (SPGM). He writes for newsletters; informally serves as an institutional historian; and edits scientific manuscripts, corporate documents, and sundry other written media. SPGM is the creative services department and hub for editing, illustration, graphic design, formatting, multimedia, and training in these areas.