Editor’s note: This is a feature on how technological changes are helping to advance science. Mention of trade names, commercial products, or organizations here does not imply endorsement by the U.S. government.
Visualizing protein structures in three dimensions instead of two has given scientists new insights into biological processes, and now artificial intelligence is adding the capacity to predict molecular behaviors that could potentially be borne out in laboratory studies.
For Kylie Walters, Ph.D., a structural biologist with the NCI Center for Cancer Research in Frederick, artificial intelligence, or AI, has revolutionized the way her laboratory works.
Walters’ laboratory specializes in structural biology—the chemistry, dynamics, and 3D structure of biological molecules. They focus on understanding pathways through which the body combats cancer naturally, mainly by removing misfolded and unneeded proteins. With the development of AI capabilities for predicting and analyzing proteins’ structures, the laboratory can use computer algorithms to hypothesize a protein’s likely structure and gain fresh insights.
“[We can] look at predicted structures [in cancer cells] and also look at predictions for protein–protein interactions and use this information to complement experimental data or to design experiments to test modeling outcomes,” said Walters.
Improved Predictions to Target Cancer-Cell Proteins
AI has proven capable of complementing experimental techniques, even surpassing them for some applications. Walters utilizes a commercial tool called AlphaFold2, an artificial intelligence program first developed in 2018 and improved in 2021.
AlphaFold2 is “trained” via a public dataset that inputs all known protein structures, as well as protein sequences with unknown structures. According to a study from Nature, AlphaFold improves its accuracy by “incorporating novel neural network architectures and training procedures based on the evolutionary, physical, and geometric constraints of protein structures.”
Walters’ team still has to test their predictions in the laboratory, but AI helps point them to a structural hypothesis for a protein they may not have experimental data on. Then, they can test the hypothesis and feed the data back to additional AI-based tools to simulate molecular behavior and better understand the structural results.
Thanks to these capabilities, Walters’ laboratory and others can better leverage molecular dynamics.
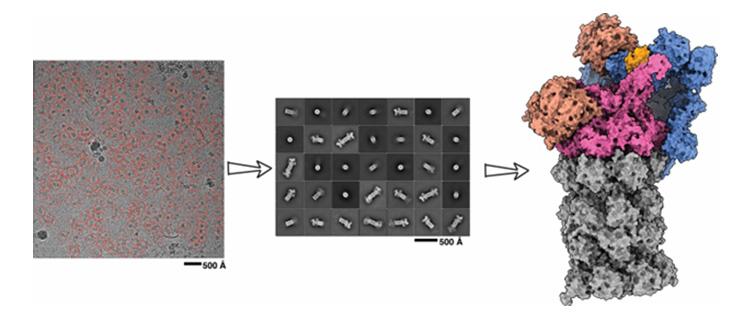
AlphaFold2 predictions also aid other experimental methods for understanding molecular structures. For example, they can serve as a starting point for cryo-electron microscopy scans, a high-resolution imaging technique used to determine the 3D structure of molecules by freezing them mid-movement. Predictions can likewise improve X-ray density maps, a tool used to determine atomic and molecular structure through another imaging technique called crystallization. In addition, AI predictions can be used to guide data analysis in nuclear magnetic resonance, a technique that uses magnetic fields and radio waves to study molecular structures.
Endless Possibilities, Realistic Considerations
While AI has already enhanced the research in her laboratory, Walters has high hopes for where it can go in the future. One development she hopes to see is an enhanced ability to predict small-molecule interactions, which will allow her team to improve the laboratory’s structure-based drug design approach. But Walters noted that for this to happen, there also has to be more laboratory data. More broadly, for AI’s value to grow, there also must be increases in funding and talent recruitment, and leaders and scientists must approach the field with an open mind.
Walters also looks forward to taking advantage of the way AI will affect the community of discovery. When scientists make discoveries, they have to figure out how they fit in the context of other discoveries made by other researchers. AI is likely to help this process in the future by capturing all relevant information and assembling it within minutes.
“I believe that these new artificial intelligence applications are going to make access to sophisticated information much easier for scientists … and the internet is going to undergo a new transformation with AI,” and “new innovation always has its adolescent periods, but the advantages and the opportunities are so much greater than the drawbacks, in my opinion,” said Walters.
Mira Deni is a Werner H. Kirsten Intern in the Partnership Development Office (PDO) at the Frederick National Laboratory for Cancer Research (FNL). In her role, she writes articles, creates communications materials, researches partners/performance measures, and more. PDO establishes partnerships with FNL and industry, academia, and other research institutions.