Many aspects of how infectious viruses assemble in cells have yet to be completely deciphered. However, as reported in a recent Journal of Virology paper, researchers may be one step closer to understanding how HIV-1, the virus that causes AIDS, assembles and replicates.
Members of the HIV Dynamics and Replication Program of the NCI Center for Cancer Research and their colleagues examined how often HIV-1 RNA molecules that reach the assembly site become part of the virus. They found that the efficiency of viral RNA incorporation into HIV-1 particles, which is necessary for HIV-1 replication to host cells, depends on the level of expression of a protein called Gag.
“During the virus assembly process, viral RNA genomes need to be incorporated into the newly formed particle to carry the genetic information to a different cell,” said Wei-Shau Hu, Ph.D., senior investigator and section head, Viral Recombination Section, Retroviral Replication Laboratory, HIV Dynamics and Replication Program, and last author of the paper.
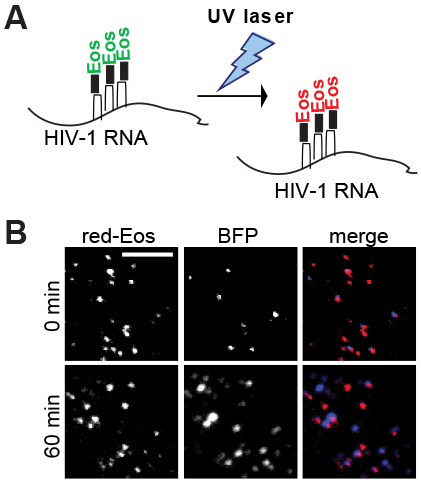
The research group studied the window of opportunity for HIV-1 viral RNA to become part of the virus. They used total internal reflection fluorescence microscopy, an optical technology for studying thin regions of specimens, to examine the interactions between viral RNA and Gag-RNA at the plasma membrane.
They found that when HIV-1 RNAs arrived at the plasma membrane and Gag was absent, the HIV-1 RNAs only stayed for a few minutes; however, when Gag was present, the HIV-1 RNAs formed complexes with Gag and could stay near the plasma membrane beyond an hour. They also found that only a portion (one-tenth to one-third) of the RNAs that reached the plasma membrane ultimately became part of the virus.
Most studies on virus assembly have used biochemical and virological approaches, Hu said. In this report, researchers used a state-of-the-art imaging method to detect RNA at single-molecule sensitivity.
“By combining [this method] with the use of a photoconvertible protein to label viral RNA, this approach allowed us to learn new insights that are not easily accessible by biochemical or virological assays,” Hu said.
First author on the paper, Luca Sardo, Ph.D., who was a visiting fellow in Hu’s lab when the paper was published, is now a researcher at the University of the Sciences in Philadelphia, where he studies aspects of HIV-1 neuropathogenesis and pharmacological latency. He earned his doctorate degree at the University of Turin, Italy.
“The advances in our understanding of HIV-1 replication have been enormous since the discovery,” Sardo said. “I firmly believe that the scientific community has never been so close to defeating HIV-1, and it is such an exciting time to be part of the global investigation that will lead to a cure.”
Hu, who earned her Ph.D. from the University of California at Davis, has worked for NCI for 16 years. Her laboratory’s long-term research interests include an understanding of how HIV transfers its genetic information during replication and how retroviruses navigate the host environment to replicate its genome.
“I believe that by studying how the virus replicates, we can identify the strengths and the weaknesses of the virus, and then design better strategies to target HIV-1,” she said.