Editor’s note: This article was originally published on the Center for Cancer Research website.
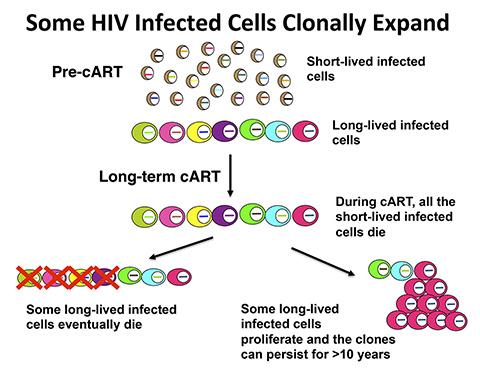
When the Human Immunodeficiency Virus (HIV) infects a cell, the virus inserts a copy of its genetic material into the host cell’s DNA. The inserted genetic material, which is also called a provirus, is used to produce new viruses. Because the viral DNA can be inserted at many sites in the host cell DNA, the site of integration marks each infected cell. Patients infected with HIV are currently treated with combined antiretroviral therapy (cART), which prevents viral replication in the majority of treated patients. When cART is initiated, most HIV-infected cells die in one or two days, and more of the infected cells die over a period of weeks to months. However there are some long-lived infected cells that do not die, which prevents patients from being cured.
Most patients are initially infected with a single HIV particle, and, as a consequence, identical viruses are found in the blood shortly after the patient is first infected. After several years of untreated viral replication, the viral sequences in the patient have diverged to the point where the viruses in the blood are all different. However, after long-term cART, identical variants of HIV are again found in the blood. Researchers are uncertain where these identical viruses come from and have suggested that, despite cART, there is some local replication in a “privileged” site or sites. An alternate hypothesis is that some of the persistently infected cells clonally expand. To investigate this idea, Stephen Hughes, Ph.D., of CCR’s HIV Drug Resistance Program (DRP), working with colleagues from the DRP, Leidos Biomedical Research, the University of Milan, the University of Pittsburgh, and Tufts University, examined provirus integration sites in samples of immune cells from five patients collected before or shortly after starting cART and after prolonged cART, ranging from five to 13 years. Because the proviruses are inserted at many places in the host DNA, integration sites can be used to identify cells that have clonally expanded from (that is, are all descendants of) a single infected parent cell.
The researchers sheared the DNA from the infected immune cells into small pieces and used polymerase chain reaction (PCR) to selectively amplify segments that contained virus-host-cell DNA junctions. Sequencing the virus-host junctions revealed the orientation and location of the proviruses. If a cell has clonally expanded, there will be more than one copy of viral DNA integrated into a specific site in the host DNA in the starting sample. When the DNA is sheared, the host DNA near the integration site is broken at different positions. Thus, if there was more than one cell with a provirus integrated at the same site in the sample, there will be DNA fragments that have the same integration site, but different host DNA breakpoints. When these fragments are amplified, the investigators can count the number of times they find the same integration site linked to different host breakpoints to estimate the number of cells in the sample that came from the same expanded clone.
As expected, before or shortly after starting cART there were many distinct viruses present in each of the five patients. After long-term cART, identical viral sequences were identified in each patient. The research team mapped 2,410 provirus integration sites, and 43 percent of these sites were from clonally expanded cells. In some cases, the clonal expansion was extensive. For example, in one patient, approximately half of the HIV-infected cells derived from a single infected cell.
Mapping revealed that there were proviruses integrated into 985 different genes. Two-thirds of the genes had one integration site and were not shown to be from clonally expanded cells. The remainder had one integrated provirus that was in a clonally expanded cell or there were multiple independent integrations in the gene, some of which were in clonally expanded cells. Of the genes with multiple independent integrations, 70 percent were genes involved in regulating cell growth. The investigators also found integrations between genes or in ambiguous sites that could not be mapped to a single site in the human genome, and some of the cells with integration sites outside of genes were also clonally expanded.
The researchers then asked whether the HIV-infected clones can persist for long periods. In one patient, they found 13 clones that were present in samples collected more than 11 years apart and another 11 clones in samples collected nearly seven years apart. Many of the persistent clones had provirus integrated into genes associated with cell growth, mitosis, or both. The scientists also found clonally expanded cells in one patient’s pre-cART sample, demonstrating that clonal expansion occurs in the absence of cART.
Finally, the investigators wondered whether provirus integration at specific sites might be associated with the clonal expansion or persistence of the infected cells. In one patient, they found 11 distinct integration sites in intron 6 of MKL2, over half of which were in clonally expanded cells, and four integration sites in intron 4 of the same gene. All 15 proviruses were in the same transcriptional orientation as MKL2. The researchers also identified an additional 15 integration sites in introns 4 and 5 of BACH2 that were also in the same transcriptional orientation as the gene. Intriguingly, both MKL2 and BACH2 are known oncogenes, genes that normally play important roles in cell growth and development but when mutated can play a role in the development of human cancer.
The scientists showed that the integration frequencies into the MKL2 and BACH2 genes were much higher in the patient samples than in two large HIV integration site libraries prepared from HeLa cells or human CD34-positive human stem cells infected in culture. Additionally, they also showed that HIV integrations in the cultured cells had no preference for specific introns nor was there any preferential integration in one orientation relative to the transcriptional direction of the target gene in the infected cell libraries. Across the entire patient data set, there was a weak, but significant, preference for proviruses to integrate opposite to gene transcriptional orientation, suggesting that integration into MKL2 and BACH2 were selected after integration, apparently because the integration altered the level of expression of the gene or led to the production of an altered protein, which in turn affected the clonal expansion or survival of the cells.
Similar mechanisms have been seen with other retroviruses that infect animals. In the animal models, insertion of a provirus in or near certain oncogenes (including BACH2 in mice) is involved in the development of cancers. Gene ontology analysis showed that integration sites in the patients’ cells, but not those in infected HeLa cells or CD34-positive stem cells, were enriched for genes in cell growth pathways. In fact, the genes identified in the patient data set were related to leukemia and Burkitt’s lymphoma while the genes identified in the data sets from cells infected in culture were not.
Together, these studies show that the persistence of expanded clones of HIV-infected cells is common and is associated with HIV integration into genes that regulate cell growth and division. These results have important implications for developing a cure for HIV—both viral replication and clonal expansion of HIV-infected cells must be prevented. The data also provide additional considerations for gene therapy applications that use retroviral vectors and raise the possibility that HIV integration may contribute to cancer development in infected individuals.
Reference
Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, and Hughes SH. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. June 26, 2014. PubMed Link.