Editor’s note: This article is part of a series about scientific devices in use at NCI at Frederick and the Frederick National Laboratory. Other parts of the series can be found here. Mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. government.
“Here are lions.” This fanciful phrase for the unknown embellishes the blank areas of antiquated maps. Sometimes cartographers used the phrase “Terra Incognita” (“unknown land”) instead. Either way, the message was clear: we don’t know what’s beyond this point.
The modern study of genetics is similar. Scientists have made great strides in studying DNA—researchers mapped the human genome in 2003—but much remains unknown, like whether or how certain mutations change biological functions or cause diseases. Moreover, most organisms’ genomes haven’t been mapped.
In short, genetic terra incognita abounds.
In cancer research, this limitation is more than an inconvenient blank spot on the map. Cancers stem from genetic mutations, changes in gene activity, and abnormal gene function. The successful development of new treatments depends in many cases on insight into these anomalies.
To address this challenge, scientists are using genetic sequencing and mapping technologies. The NCI Center for Cancer Research Sequencing Facility (operated by the Frederick National Laboratory) is one of many groups exploring the unknown. Here, a team of genetic experts—modern-day molecular cartographers—employ top-of-the-line equipment to help scientific collaborators investigate DNA, RNA, and a myriad of sequencing questions.
A Small Approach to Answer Big Questions
The Illumina NovaSeq 6000 is one of these devices. It’s a platform for short-read sequencing, a method for understanding short DNA strands by determining the order of their nucleotide bases—the A, C, G, and T that comprise genes. With it, scientists can visualize unknown genes or discover mutations, like incorrect bases and sequences, in known ones.
Before being loaded into the NovaSeq, millions of copies of a single-strand DNA section are placed onto a small glass chip alongside complementary molecules and free-floating nucleotide bases tagged with fluorescent particles. Once inside, a chemical reaction begins. The molecules connect to the DNA strands and attract the free-floating bases, which attach to the molecule one by one, pairing up with the complementary base on the strand. Each base’s fluorescent tag breaks off, creating a small burst of color-coded light that’s captured by the NovaSeq’s camera.
Data from the lights tells scientists which bases connected to the sequenced strand and, by extension, which bases are on the strand. For instance, if the color indicates that guanine (G) connected, its partner, cytosine (C), is on the sequenced strand. Sequencing software combines the data, hundreds of millions of snippets of sequence, to map the strand.
On the NovaSeq’s largest setting, the Sequencing Facility staff can fully sequence 48 human genomes in just two days, generating terabytes of data in the process. The platform is also flexible enough to sequence old or lower-quality DNA samples.
“Things that have been sitting in the repository or freezer for 30, 40 years, sometimes we are able to retrieve them and do some wonderful work,” said Jyoti Shetty, head of the Sequencing Facility’s Short-Read Technology Section.
However, the NovaSeq faces the same challenge as many short-read platforms: it sequences diminutive DNA fragments, as short as 75–600 base pairs, but most genomes are millions or billions of pairs long and contain many repeated regions. That makes it difficult to accurately assemble larger maps.
A Wider View
Another device at the Sequencing Facility, the Pacific Biosciences (PacBio) Sequel, helps to fill this gap. As a platform for long-read sequencing, it allows the team to study much longer fragments—DNA and RNA strands 250–30,000 base pairs long.
“You’re not going to get hundreds of millions of reads like you do on Illumina, but you get longer read lengths, which is a good thing, especially if you’re doing genome assembly,” said Chris Lyons, acting manager of the Sequencing Facility’s PacBio Operations Section, adding that longer reads make it easier to recognize and assemble repeated or challenging regions.
The Sequel functions similarly to the NovaSeq, using a polymerase molecule that attracts fluorescently tagged bases. Copies of a DNA or RNA strand are loaded onto a chip containing one million microscopic wells called zero-mode waveguides. Inside the machine, the polymerases run alongside the strands and attract the bases, like a person’s finger sliding along words on the page of a book. The Sequel shines a light into each well, photographing the fluorescent bursts with its camera.
Although the Sequel enables more accurate genome assembly, it too has limitations. It requires large amounts of high-quality genetic samples, and neither it nor the NovaSeq can investigate larger genetic questions, like chromosome-scale mutations that are millions of base pairs long.
The Big Picture
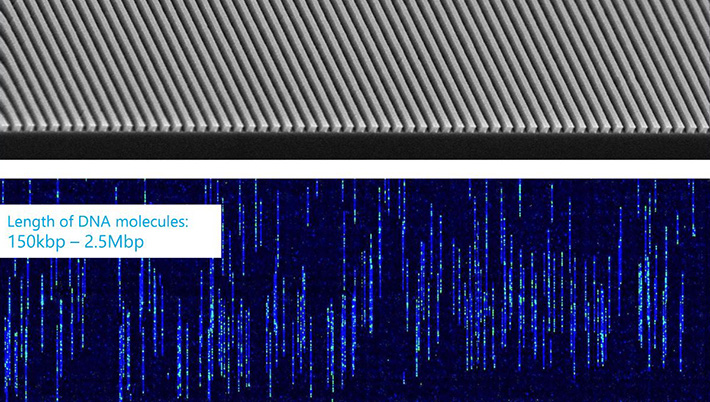
For such tasks, the Sequencing Facility turns to the Bionano Genomics Saphyr and optical mapping, a technique for assembling the genome by measuring the distance between marked sections of DNA. The DNA molecules examined with the Saphyr are massive, hundreds of thousands to millions of base pairs long.
The DNA is first introduced to fluorescent tags that are chemically programmed to attach to certain genetic sequences. After the DNA fragments are placed into the Saphyr, they are untangled, straightened, stretched, and sieved through narrow channels. A camera captures the fluorescence as the strands pass through, creating a color-coded map. Thanks to the chemically programmed attachment, if a certain color appears, the scientists will know which sequence occupies that location on the strand.
Because the DNA segments are so large, using the Saphyr to map a complete chromosome is like assembling “a ten-piece puzzle,” says Monika Mehta, Ph.D., head of the Sequencing Facility’s Research and Development Section. In comparison, using the NovaSeq or Sequel is like assembling “a thousand-piece puzzle.”
To put it another way, if the human genome were the size of the United States, the NovaSeq could map one house, the Sequel could map a few streets, and the Saphyr could map an entire state. However, unlike the other two, the Saphyr doesn’t provide many details on smaller sections of the genome.
“So we can combine these two data sets: we can take the sequence that we have generated with either Illumina or PacBio; we can combine it with the optical maps that we generate, and that can give us chromosome-length information,” Mehta said.
The three devices complement each other, overcoming limitations and amplifying strengths when used together. To help with such projects, the Sequencing Facility’s Bioinformatics Group has even developed custom-built pipelines that integrate the massive amounts of data for analysis. The facility’s collaborators rarely need that much detail for a single project, but Shetty says those opportunities are “really exciting.”
In a recent collaboration with the Frederick National Laboratory’s Protein Expression Laboratory, the Sequencing Facility combined the methods to assemble the sequence of a cell line that’s derived from cabbage looper moths and commonly used to produce recombinant proteins. The facility generated the long-read sequence data, combined it with short-read sequence data, and integrated them with optical maps. The resulting high-quality genome assembly will enable improvements in protein production using the cells.
With such an array of tools at its disposal, the facility is helping to push back the genetic terra incognita project by project, erasing the lions and filling in the blank spots on the map.
The Sequencing Facility and its devices do more than DNA sequencing. For more information about its services, contact Bao Tran, director of the Sequencing Facility, at Bao.Tran@nih.gov.