Steve Hughes, Ph.D., compares HIV research to a war. He and his colleagues are entrenched on a microscopic battlefield, fighting a conflict where seemingly small victories could mean a leap forward.
Right now, they are grappling with the emergence of HIV strains resistant to existing antiretroviral drugs, medicines that suppress the virus in people living with HIV. The team’s current target is integrase, an enzyme that HIV uses to insert a DNA copy of its genetic information into the DNA of a host cell, thereby allowing the virus to replicate and spread.
HIV research and treatment have come far in the four decades since the virus was discovered. Today, most infections can be managed by therapies that use a combination of state-of-the-art antiretroviral drugs. But mutated strains that resist even the best HIV drugs can arise. Developing treatments for them is an ongoing battle in a long war.
“There are some mutants that are a problem, and the ones that are really bad are a problem for everybody,” said Hughes, chief of the Retroviral Replication Laboratory and senior investigator in the HIV Dynamics and Replication Program at NCI at Frederick.
Even small changes in HIV can cause substantial resistance. An alteration to one or two of the key amino acids in integrase (amino acids are the “building blocks” of proteins, including enzymes like integrase) is enough for the virus to become resistant to integrase strand transfer inhibitors (INSTIs), the class of antiretrovirals that block the integration of viral DNA.
Battle Plans
Hughes and his colleagues are working to develop INSTIs that can defeat most, or all, of the resistant viruses. Given that resistant HIV strains are on the rise, the stakes are understandably high.
“In this war, there is a conflict between the viral enzyme—integrase—which is trying to figure out clever ways to reject the inhibitor but retain the ability to bind its normal substrates, and we’re trying to find ways to make that as hard as possible,” Hughes said.
It’s a complex problem. Hughes says the key is to get ahead of the virus—to imagine what could go wrong and launch a preemptive strike before the situation deteriorates.
“When I started on this problem … [HIV] was a death sentence,” he said, recalling the early research in the 1980s. “If we lose the game against drug resistance, we’re going to be back where we were when the virus was first discovered.”
Scientists from multiple fields have partnered to find and target weaknesses in integrase. Hughes’ team, experts in HIV biology, work with Terry Burke, Ph.D., and chemists in the Chemical Biology Laboratory at NCI at Frederick to design and develop new INSTIs. Robert Craigie, Ph.D., and his team at the National Institute of Diabetes and Digestive and Kidney Diseases engineer and purify versions of integrase for biochemical analysis and structural studies.
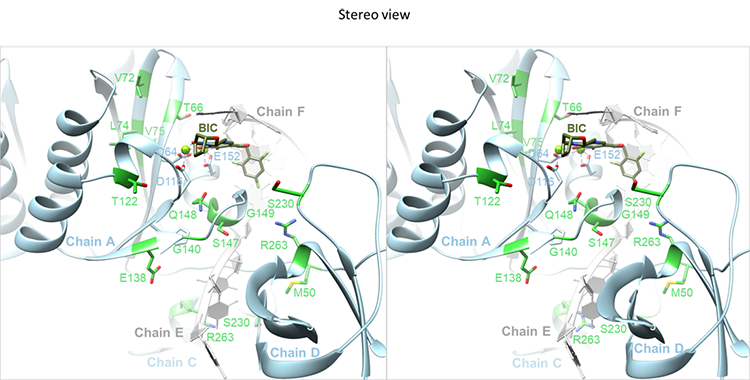
They’re joined by Dmitry Lyumkis, Ph.D., whose structural biology laboratory at the Salk Institute is using electron microscopy to solve the structures of the INSTIs bound to HIV integrase at atomic resolution, obtaining high-quality images of the interaction.
Microscopic Maneuvers
Structural biology is an important weapon in the war. By understanding how INSTIs bind to integrase at the atomic level and how changes in the drug-resistant mutants interfere with the binding, the team can design better compounds.
“These structures are epiphanies. The details of the binding of INSTIs to HIV integrase are the interactions that save people’s lives made visible,” Hughes said. “It’s magic to be one of the first humans on the planet see something like that.”
Earlier this year, the team published the first structures of HIV integrase bound to INSTIs. With that crucial data now in hand, they will turn their attention to the structures of drug-resistant mutants.
Unfortunately, there are hundreds of mutants and dozens of INSTIs—too many possible combinations to image them all. To narrow down the field, Steve Smith, Ph.D., a research biologist in the Hughes laboratory, recently led a study to identify the most problematic mutants. He tested how both FDA-approved INSTIs and experimental INSTIs developed by the Hughes and Burke laboratories performed against the worst mutants. The work pinpointed two INSTIs, including one designed at NCI at Frederick, that are particularly promising—but it also showed that some mutants cause problems for all known INSTIs.
Craigie’s team is engineering and preparing versions of the worst mutant integrases for structural analysis. The enzymes and the promising compounds will be sent off to Lyumkis, who will solve their structures.
“We’re in the process of trying to better understand the physical interactions between the inhibitors and … HIV integrase. And if we can come to grips with that, we hope to use that information to plan more effectively, and more efficiently, how to make the right kinds of inhibitors,” Hughes said.
Hughes is confident this approach will be more efficient than the traditional approach, used when structural information isn’t available: blindly creating a huge series of compounds in hopes of finding one that works. He also recognizes that the partnership isn’t fighting the battle alone. There are other teams of scientists striving to defeat drug-resistant integrases via similar methods, and he believes in advancing toward a solution together.
“We want to make sure, as much as possible, that the prescriptions for how to create better INSTIs are available to everyone so that everyone has the opportunity to succeed. We certainly want to get credit for what we do, … but we want to make the information public,” he said.
“The most important thing is to win against HIV, not against our competition in science.”