NCI at Frederick’s latest class of student interns continues to telework under pandemic protocols. While that means not working in laboratories or offices as they would in a normal year, it doesn’t mean a lack of opportunities to make a difference in science.
In fact, according to Kedar Narayan, Ph.D., volume electron microscopy group leader at the Center for Molecular Microscopy, one opportunity is quite unique.
The project is called Mito Mega Mix, the brainchild of Narayan and Marsha Nelson-Duncan, program manager of education, outreach, and training at NCI at Frederick. Participating interns become teachers who help to train an artificial intelligence (AI) algorithm to recognize mitochondria in electron microscopy images.
“You’re part of something that hasn’t been attempted before in this manner,” Narayan told the interns. The crowdsourced approach allows the AI to benefit from diverse perspectives, which enriches the data and makes the algorithm more robust.
The opportunity to help is opening to a larger crowd, too. Any student intern at the National Institutes of Health is now welcome to participate.
Learning by Repetition
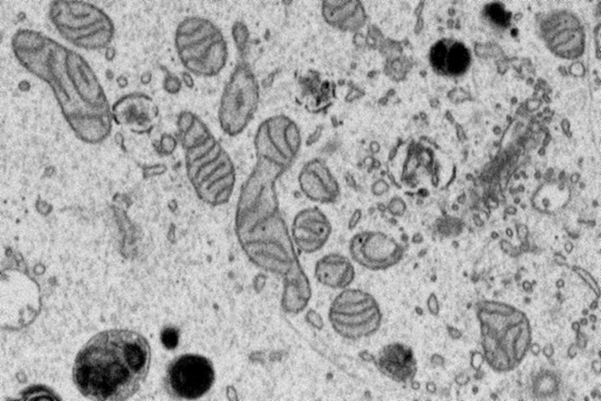
Mitochondria are cellular structures that convert nutrients to energy. They’re important targets for study: visualizing their shapes through electron microscopy can help scientists better understand several diseases.
Unfortunately, identifying mitochondria segments manually in the thousands of 2D grayscale images created by an electron microscopy scan “is really very difficult,” Narayan said. Assembling those images into a 3D structure of a complete mitochondria is even harder. AI algorithms can do it faster and more efficiently than humans, but they need help to be effective.
“Our AI algorithms can be thought of as algorithms that learn concepts—if you think about it, a mitochondrion is a pretty complex concept! But they need data to learn from, and in our approach, we humans help train them,” Narayan said.
Mito Mega Mix is an attempt to fill the gap. It takes place on a web-based platform, where Narayan and his team curated and uploaded thousands of microscopy images. Some contain mitochondria. Some are devoid of them. Participants use the platform’s pen tool to trace the mitochondria in the images. Narayan and his team feed those annotated images to the algorithm for analysis. He said it’s similar to teaching a baby to recognize shapes.
The AI becomes better at recognizing mitochondria through repetition, by analyzing more annotated images. Eventually, it will be powerful enough to do the annotation independently, the goal of the project.
Collaboration, Consequential Science, and a Bit of Competition
For participants, Mito Mega Mix is more than a scientific “assembly line” or mountains of markup work. They’re contributing to the eventual scientific publication about the algorithm.
They’re also generating data and making meaningful decisions. The platform requires participants to rate the difficulty of identifying mitochondria in each image. That data helps Narayan’s team calibrate the algorithm for difficulty, which increases its power further.
The project represents an educational and collaborative opportunity, especially for interns. The website is brimming with tools, background information, and comprehensive guides to help participants. A discussion board offers a means to ask questions and work through challenges with others.
“The … project is a fun and relaxing activity that has allowed me to identify the variety of looks that a mitochondrion has in images captured by electron microscopes,” said intern Jiayi Wang.
It’s not the same as working together in laboratories, but the setup tries to capture the spirit of it as closely as possible. For interns, it makes the socially distanced experience more engaging and scientifically valuable.
“This [project] is a first for us. We’re very excited,” Narayan said. “We have a stellar group of kids.”
Narayan and Nelson-Duncan have also sprinkled some friendly competition into the collaboration. Participants are scored on accuracy and the number of images completed, and the Mito Mega Mix home page displays the names of the top participants. Nelson-Duncan is awarding prizes to the leading interns.
Mito Mega Mix can be accessed from any computer or tablet. National Institutes of Health student interns interested in learning more about the project should contact Narayan at kedar.narayan@nih.gov. Those ready to participate should contact Nelson-Duncan at marsha.nelson-duncan@nih.gov to be set up with an account.
“We want you to do real science,” Narayan told the interns. “We want you to be a part of what we do here.”
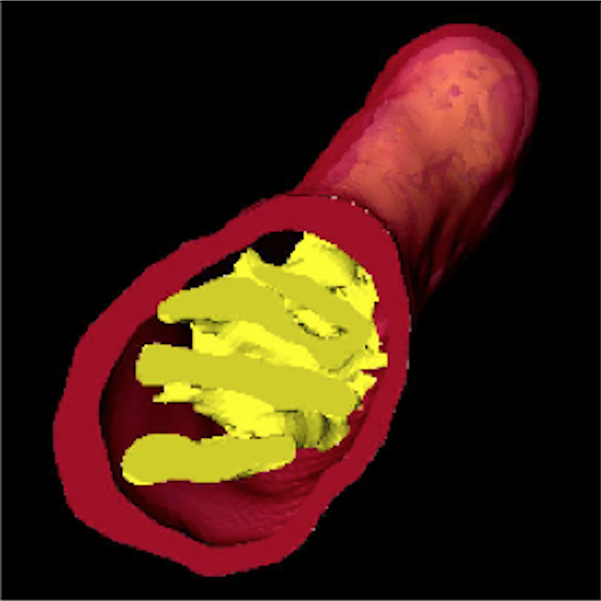
Video: A 3D projection of a mitochondrion assembled from a segmented stack of 2D grayscale electron microscopy images.
Samuel Lopez is a technical editor in Scientific Publications, Graphics & Media (SPGM), where he writes for NCI at Frederick and Frederick National Laboratory’s news outlets; manages the day-to-day operations of the Poster newsletter; informally serves as an institutional historian; and edits scientific manuscripts, corporate documentation, and a slew of other written media. SPGM is the facilities’ creative services department and hub for editing, illustration, graphic design, formatting, and multimedia training and support.