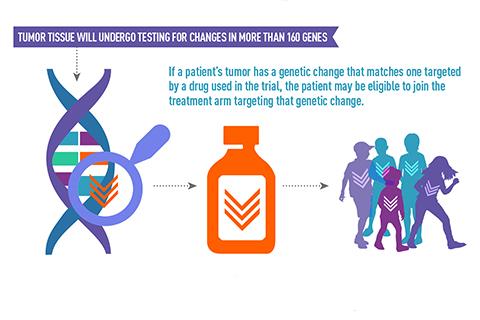
The National Cancer Institute and Children’s Oncology Group recently opened enrollment for a new Phase II trial of personalized precision cancer therapies. Called the Pediatric Molecular Analysis for Therapy Choice (Pediatric MATCH), the trial seeks to treat children and adolescents aged 1–21 whose solid tumors have failed to respond to or re-emerged after traditional cancer treatments.
The goal of Pediatric MATCH is to pair mutations in children’s tumors with experimental drugs that target those mutations. Participants will have tumor specimens sequenced on a targeted next-generation sequencing assay developed for use in this study. The Children’s Oncology Group will then upload the resulting data to a custom database called PED-MATCHBox that will evaluate the tumors and, if the specimen tests positive for a targetable mutation, pair it with the corresponding drug.
The hope is that, by pairing a mutation with a drug that is specifically designed to target that mutation, patients will see improved responses to therapy and increased survival rates. The resulting data will also allow researchers to better evaluate drug efficacy and plan future treatments.
Pediatric MATCH is an outgrowth of NCI-MATCH, an ongoing precision oncology medicine trial in adults that was created by NCI and supported by the Frederick National Laboratory for Cancer Research (FNLCR). Early returns from NCI-MATCH have been used to improve the model for Pediatric MATCH. The NCI-MATCH assay began by sequencing 143 specific genes, but subsequent advancements have allowed the Pediatric MATCH sequencing assay to start with 161 genes. Similarly, the information gained from NCI-MATCHBox—PED-MATCHBox’s older counterpart—was used to develop its more powerful successor.
Tony Fu, deputy director, Biomedical Informatics Applications Development Group, Data Science and Information Technology Program, FNLCR, says that although the informatics system for both NCI-MATCH and Pediatric MATCH uses the same knowledge-based AI core to match patients to experimental drugs, PED-MATCHBox represents a significant improvement.
“We have taken the lesson learned from NCI-MATCH and built a more secure, performant, flexible, and robust informatics system in the Amazon Web Services cloud environment to support the Pediatric MATCH Phase II clinical trial,” Fu said.
Fu, along with his colleague Brent Coffey, director of FNLCR’s Biomedical Informatics Application Group, was among the collaborators from FNLCR and NCI’s Center for Biomedical Informatics and Information Technology who created PED-MATCHBox and NCI-MATCHBox. In preparation for Pediatric MATCH, the Center provided software architecture and best-practices support to FNLCR’s team as it built the database system. The Center was also instrumental in uploading the database to the cloud environment.
“[We built a] completely new system from the ground up,” said Coffey. “The collaboration between people who worked on the adult study has led to a more robust informatics system, more advanced artificial intelligence, and more flexibility of the code base to evolve as the needs of the trial change.”
Since its launch in 2015, NCI-MATCHBox has evaluated data from over 6,100 patients and analyzed over 5,800 samples using an initial set of rules created for drug assignment. PED-MATCHBox will expand those initial rules, allowing for quicker, more precise assignment.
“In many ways, Pediatric MATCH is a more complex study than NCI-MATCH,” said David Patton, director of the Federal Informatics Program and informatics chair for MATCH, Center for Biomedical Informatics and Information Technology, NCI, who has worked closely with Coffey’s FNLCR team for both NCI-MATCH and Pediatric MATCH. “The team not only applied knowledge acquired during NCI-MATCH, they leveraged innovative approaches in addressing the unique challenges presented by this novel pediatric study design. I am very impressed with the final product delivered by the entire Pediatric MATCH team.”
FNLCR has made other crucial contributions to both Pediatric MATCH and NCI-MATCH, as well. The staff of the Molecular Characterization Laboratory, led by Robin Harrington, David Sims, and Anand Datta, Ph.D., created the single-sequencing assay used in the two trials, and they have sequenced more than 1,000 tumor biopsies taken as part of NCI-MATCH.
To sequence the tumor samples for Pediatric MATCH, the Molecular Characterization Lab will work alongside the University of Texas MD Anderson Cancer Center. The goal is a sequencing turnaround time of 14 days, which would allow for rapid uploads to PED-MATCHbox that in turn enable faster tumor–drug pairing and more efficient treatment.
“Being able to play a role in both of the MATCH trials has been extremely gratifying for our team, knowing we are contributing to the testing of new and promising precision medicine treatments,” said Mickey Williams, Ph.D., director, Molecular Characterization Laboratory. “These studies are open in many hospitals in the United States, and many patients have been treated in their own community.”
The trial is projected to enroll 200–300 patients per year at approximately 200 nationwide sites and hospitals managed by the Children’s Oncology Group, a member of NCI’s National Clinical Trials Network. Researchers have already begun sub-studies testing seven experimental drugs, and they expect to add more drugs and sub-studies as the trial progresses.
Those involved have high hopes for Pediatric MATCH, and they see it as an example of how intramural and extramural collaboration can move cancer research forward.
“[We] don’t have results yet, as it’s a multi-year study,” Coffey said. “But I would say that this is a large project that is possible because of a diverse set of skills coming together.”
Fu agreed and added, “We hope that treating individual genetic abnormalities will one day be the standard of care and that this approach will alleviate suffering and save the lives of many people.”