![]() Editor's note: Platinum Highlight articles are noteworthy publications selected periodically by Craig Reynolds, Ph.D., associate director, NCI at Frederick, from among the most recently published Platinum Publications.
Editor's note: Platinum Highlight articles are noteworthy publications selected periodically by Craig Reynolds, Ph.D., associate director, NCI at Frederick, from among the most recently published Platinum Publications.
Until recently, researchers studying RAS, a family of proteins involved in transmitting signals within cells, believed that the exchange of guanosine 5’-diphosphate (GDP) by guanosine triphosphate (GTP) was sufficient to activate the protein. Once activated, RAS can cause unintended and overactive signaling in cells, which can lead to cell division and, ultimately, cancer.
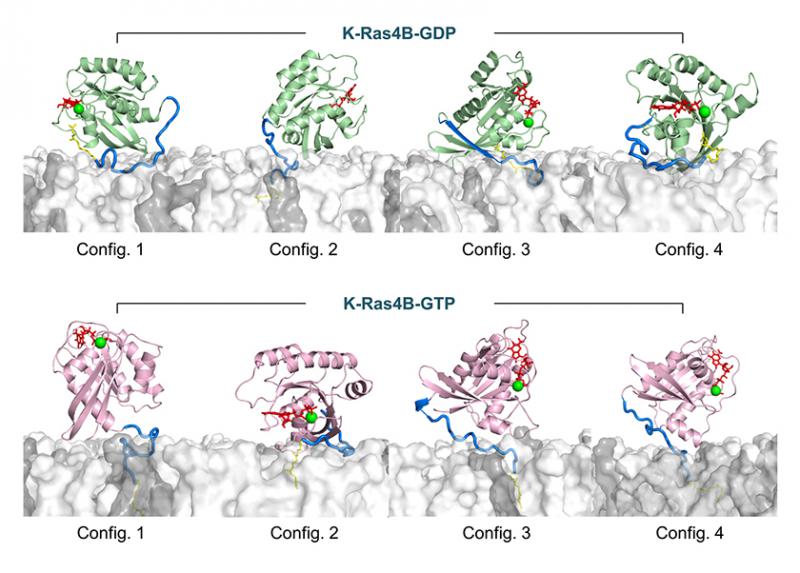
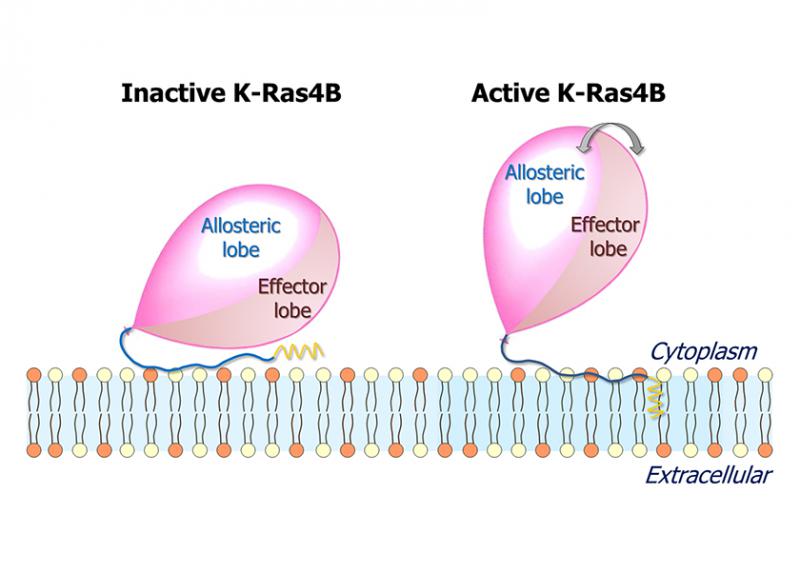
However, as reported in a FASEB Journal paper, the GDP/GTP exchange may not be sufficient for activation. Instead, the results published by Hyunbum Jang, Ph.D., Ruth Nussinov, Ph.D., and their colleagues suggest that a number of factors, including the GDP/GTP exchange, hypervariable region sequestration, farnesyl insertion, and orientation/localization of the catalytic domain at the membrane conjointly determine the active or inactive state of K-Ras4B, the member of the RAS family on which the study focused.
“K-Ras4B is the most oncogenic isoform of the RAS family,” Nussinov said, meaning it is the most active contributor to tumor formation. “Clarifying the mechanism of K-Ras4B activation and identifying the active conformation are important goals and will be useful in rational drug discovery.”
According to Nussinov, the team chose to study RAS activation because the molecular biophysics of the protein—its environment and signaling, from a structural standpoint—can help explain the Ras mechanism in oncogenic cells and help predict drug resistance.
Previously, the team had published a semisynthetic method for production of farnesylated and methylated K-Ras4B, and a modified version of that method was used in the research discussed here. To obtain structural details of the farnesylated K-Ras4B, the team performed molecular dynamics simulations on K-Ras4B both in aqueous and lipid environments. Their observations led them to conclude that GTP binding alone does not determine an active conformation.
Jang, the first author on the paper, said “This is the first computational effort to model the full-length K-Ras4B protein with the post-translational modifications interacting with the PS-rich membrane.”
The research group completed the computational part of the study at the NCI at Frederick campus using the Biowulf high-performance computing system. The experimental nuclear magnetic resonance study was performed in the University of Illinois in Chicago.
Nussinov has worked at the NCI at Frederick since 1985, while Jang has worked there since 2005.