A recent Data Science Discussion Panel, hosted virtually by the Scientific Library, provided more than 60 participants with expert insights about data science in cancer research from six members of Advanced Biomedical Computational Science (ABCS), part of the Biomedical Informatics and Data Science Directorate of the Frederick National Laboratory for Cancer Research.
ABCS supports scientific research at the Frederick National Laboratory for Cancer Research, the National Cancer Institute, the National Institutes of Health, and other federal agencies. Because bioinformatics—one of the drivers of modern life sciences—is a field that has cross-disciplinary uses, it’s important to foster multi-laboratory collaborations that can help tackle complex scientific challenges. As the Scientific Library is aware of the breadth of research across many federal laboratories in Frederick, it encourages scientific interaction and collaboration and, therefore, hosted a discussion panel to provide a platform where data science experts could share their expertise with other scientists. This discussion panel is the first in a series to enrich biomedical research and strengthen research support services at NCI at Frederick.
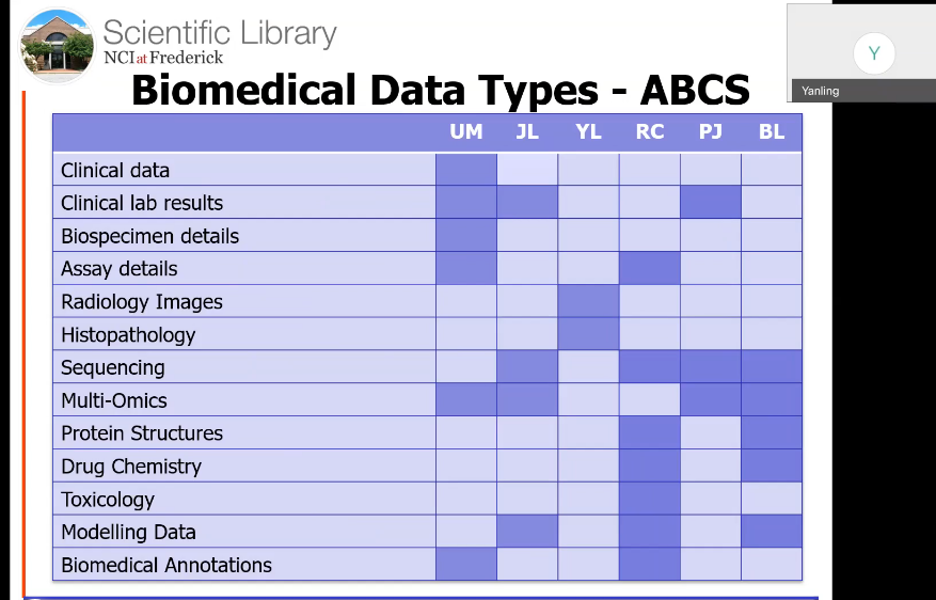
Panelists included Uma S. Mudunuri, deputy director and data solutions team lead; Justin B. Lack, Ph.D., NIAID bioinformatics team lead; Yanling Liu, Ph.D., imaging and visualization team lead; Raul Cachau, Ph.D., senior principal scientist; Parthav Jailwala, CCR bioinformatics team lead; and Brian T. Luke, Ph.D., principal scientist.
Mudunuri started the panel with a brief overview of the steps for understanding, processing, extracting value from, visualizing, and communicating research data. “[We are] trimming down those billions and trillions of data points to a one- or two-page report before [we] communicate it to different audiences,” she said.
The six panelists then discussed the many types of research data they work with, ranging from clinical data and radiology images to assay details, sequencing, multi-omics, and protein structures. According to Mudunuri, “For any users of the Frederick cluster, we do maintain all of the bioinformatics databases and software. These are hundreds of databases, so we have applications covering genes, proteins, variants, drugs, diseases, and other biomedical databases.”
To speed up the analysis process for the diverse range of data, the group develops automated workflows each time they encounter a novel data type, Lack said.
Next, the panelists described some challenges they commonly face when working with research data and their solutions. Luke explained that data analysts need to understand the science behind the data before they can effectively analyze and communicate it. “You have to understand what’s going on in an experiment in order to understand what the data really means,” he said.
The panelists continued the discussion by sharing their thoughts on the benefits, drawbacks, and obstacles to data sharing in biomedical research. Mudunuri described a lack of clarity in data sharing due to the large number of data repositories that have to be searched individually and the lack of guidelines on where different types of data sets should be shared. She mentioned that the NIH Genomic Data Sharing Policy has helped with defining how genomic data sets can be shared. She also described resources like Google Dataset Search, policies from the NIH Office of Science Policy, and NCI initiatives, which streamline the processes and policies for searching and sharing data sets.
The panelists concluded the discussion by providing advice on best practices for working with biomedical research data. Mudunuri shared that the most important thing to remember for any data-science-related project is not to make any assumptions in the data science process. Lack advised researchers to have a thorough analysis plan in place before any data is generated. He also stressed the importance of exploring and reviewing the data for quality-control purposes, since what may seem to be an interesting result may actually be driven by some technical artifact or problem with the data.
Jailwala and Liu both advised that researchers contact ABCS early in the research process, before the data is generated so that ABCS can assist with experimental planning and design. Luke encouraged researchers hoping to learn more about data analysis to gain experience in as many areas as they can since they may need the knowledge later. Luke also noted that researchers shouldn’t assume causality if the data appears to show a correlation and that results should be tested further. Cachau cautioned that sometimes the information sought may not be in the data.
During the question-and-answer session, a participant asked Mudunuri to comment on her team’s work with aggregating clinical data. Mudunuri explained that data sets from different clinical sources can vary greatly, so integration requires a lot of quality assurance and standardization.
Another participant asked about the best avenues for sharing data science pipelines with intramural and extramural collaborators. Liu and Jailwala described their experiences with data-sharing tools accessible through websites developed by ABCS and the release of end-to-end pipelines developed by ABCS.
Researchers can submit requests for experimental design consultations through the project request form on the ABCS website.
Future Discussion Panels
The Scientific Library provides a virtual space for scientists to learn from one another. Due to the success of the Data Science Discussion Panel, the Scientific Library will be hosting more virtual discussion panels where participants can learn about services to support their research.
On September 16, from 2:00–3:00 p.m., members of the Scientific Publications, Graphics and Media group will discuss their process, tools, challenges, and best practices for editing, formatting, and illustrating scientific publications. Access the discussion panel via WebEx.
If your team provides services to NCI at Frederick and Frederick National Laboratory researchers and staff and is interested in sharing its expertise, contact Joelle Mornini to participate in a discussion panel.