Susan Lea, D.Phil., and her team of microscopists at NCI Frederick have a mountain of data on their hands.
Their microscope, a state-of-the-art Titan Krios G4, collects an average of 5 terabytes of data daily. That’s roughly equivalent to storing 1,250 movies or 2.4 million e-books.
It would be unwieldy if it weren’t for a partnership with the Enterprise Information Technology Directorate (EIT) at Frederick National Laboratory. Thanks to their support, Lea and her team in the Center for Structural Biology have all the capacity they need to manage the data in-house.
It’s hard to overstate the impact. Uploading the multitudinous data files somewhere for analysis—even an accessible place like the National Institutes of Health’s powerful Biowulf computing cluster in Bethesda—is off the table. Any sort of useful science would be impossible.
“[I’d] just be sitting there all the time waiting for my multiple terabytes of data to copy from one place to another so I could do something to them,” said Lea, Center for Structural Biology chief.
Scanning the Structures
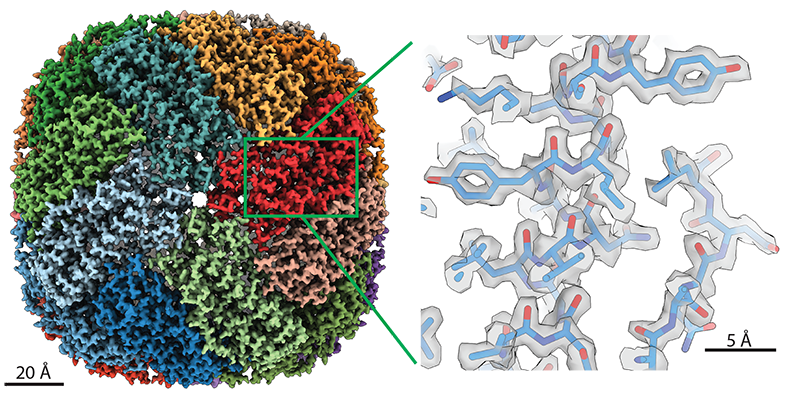
The data come from the thousands of images the Krios collects during microscopy scans. Lea’s team, part of the Center for Cancer Research, uses the instrument for cryo-electron microscopy (cryo-EM). Through rigorous computing and analysis, the method assembles the 2D images into a high-resolution 3D structure of the subject. These structures help scientists understand the biological function of the molecules they’re studying.
Cryo-EM is even more detailed than X-ray crystallography—a popular structural imaging technique—and can image harder targets. Under optimal conditions, Lea’s Krios is capable of capturing individual atoms.
With cryo-EM, Lea’s lab focuses on scanning molecular interactions and large molecules. In cancer and disease research, where it’s helpful to know how drugs connect with their targets, the ability to see these interactions is tremendously powerful.
Making It Manageable
Jay Knight, IT Operations Group director, has been leading EIT’s efforts to build and maintain the computing structure to make that possible for two years now. To accommodate the first round of upgrades, the project began eight months before Lea and her laboratory even came to Frederick.
Over the partnership, Knight’s expert team has installed new, tailored storage capabilities on Frederick’s computing network. Like swapping a small funnel and container for larger ones, they increased the capacity for data upload so Lea’s laboratory can properly move and store data. Firewall adjustments ensured the data stayed secure and enabled the Krios to connect to the network.
EIT has also reconfigured capabilities for the microscopy analyses. Frederick’s own newly resurrected high-performance computing cluster—Biowulf’s little cousin—received an upgrade.
The upshot is stronger computing power across Frederick, Knight said. His team has also learned from the process and is equipped to apply similar techniques to other projects.
“The computing [team] have been great, and they've been really interested in establishing a collaboration. I think that's what you need,” Lea said. “This isn't trivial computing to set up, and so we needed IT specialists who are interested in making it … work.”
Positioned for Success
Important contributions have come from others, too. Hans Elmlund, Ph.D., a senior investigator in the Center for Structural Biology, and his team designed the algorithm that analyzes the data from the Krios.
Combined with EIT’s work, that’s made it possible for the Krios to upload its data and images to the network while the hours-long microscopy scan is underway. This lets the computer analyze them even as the rest of the scan finishes.
“[You can] generate the high-resolution 3D reconstruction straight after data collection is done,” Elmlund said. “We are in the forefront when it comes to this stream-processing aspect.”
NCI Frederick and Frederick National Laboratory offer an uncommon opportunity to unite science, technology, and technical support to this degree. The large research portfolio and diversity of expertise, coupled with public funding, gives staff room to adopt new methods, devise creative solutions, and investigate scientific questions that other institutions can’t.
In situations like Lea and Knight’s partnership, the benefits are twofold. Not only does Lea’s team have the support to do meaningful research, the technical advances help other scientists. More computational work and analyses can now occur in Frederick rather than through Bethesda’s Biowulf or elsewhere, saving time and effort.
“Based on our experience with Dr. Lea, … we would like to take that model and see if we can bring it out to the rest of the [Frederick] scientific community, start more partnerships,” Knight said. “We’re here to help.”
Editor’s note: A version of this article originally appeared on the Frederick National Laboratory website under the title, “Computing collaboration makes massive microscopy analyses possible.” Mention of trade names, commercial products, or organizations here does not imply endorsement by the U.S. government.
Samuel Lopez leads the editorial team in Scientific Publications, Graphics & Media (SPGM). He writes for newsletters; informally serves as an institutional historian; and edits scientific manuscripts, corporate documentation, and a slew of other written media. SPGM is the creative services department and hub for editing, illustration, graphic design, formatting, multimedia, and training in these areas.